These lectures also cover UNIXLinux commands and some programming elements of R a popular freely available statistical software. A set of lectures in the Deep Sequencing Data Processing and Analysis module will cover the basic steps and popular pipelines to analyze RNA-seq and ChIP-seq data going from the raw data to gene lists to figures.
Incorporating Rnaseq Tophat To Augustus Computational Biology
One of the most popular RNA-seq mappers is TopHat which aligns reads in two steps.
. MRNA rRNA miRNA converting in some way to DNA and sequencing on a massively parallel sequencing technology such as Illumina Hiseq. Much of Galaxy-related features described in this section have been developed by. RNA-seq as a genomics application is essentially the process of collecting RNA of any type.
RNA-seq transcriptome sequencing is a very powerful method for transcriptomic studies that enables quantification of transcript levels as well as discovery of novel transcripts and transcript isoforms. One of CBSU BioHPC Lab workstations has been allocated for your workshop exercise. This tutorial is inspired by an exceptional RNA seq course at the Weill Cornell Medical College compiled by Friederike Dündar Luce Skrabanek and Paul Zumbo and by tutorials produced by Björn Grüning bgruening for Freiburg Galaxy instance.
If you have Bowtie 2 installed and want to use it with Tophat v20 or later you must create Bowtie 2 indexes for your. We recommend that you watch the video Aligning RNA-seq reads to reference genome instead which covers t. This practical will introduce some popular tools for basic processing of RNA-seq data.
TopHat2 uses using Bowtie to align RNA-Seq reads to mammalian-sized genomes and then analyzes the mapping results to identify splice junctions between exons. To find junctions with TopHat youll first need to install a Bowtie index for the organism in your RNA-Seq experiment. TopHat is a collaborative effort between the University of Maryland Center for Bioinformatics and Computational Biology and the University of California Berkeley Departments of Mathematics.
This tutorial will focus on doing a 2 condition 1 replicate transcriptome analysis in mouse. The allocations are listed on the workshop exercise web page. Bowtie1 is an ultrafast memory-effi cient aligner for large sets of short reads.
Classroom Guide for Students. Using TophatCufflinks to analyze RNAseq data. Tophat rna seq tutorial Written By kendrickbrenner11517 Thursday March 31 2022 Add Comment Edit.
Everyone should have a BioHPC account to access the computer. To install TopHat from source package unpack the tarball and change directory to the package directory as follows. Find out the name of the computer that has been reserved for you httpscbsutccornelleduwwmachinesaspxi88.
RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Hands-on using TopHat and Cufflinks in Galaxy Tutorial 3 Advanced RNA-Seq Analysis topics. Prepare the working directory. Httpcbsutccornelleduww1sessionaspxwid9sid12 Please consult the PDF file with instructions on how to access and use the Lab workstations for the.
The user ID is normally your Cornell NetID. It aligns RNA-Seq reads to mammalian-sized genomes using the ultra high-throughput short read aligner Bowtie included in this plugin and then analyzes the mapping results to identify splice junctions between exonsThis plugin runs on Mac OS and 64-bit Linux only it is not supported Windows. In this tutorial well map reads from an RNA-seq study in Drosophila melanogaster to the reference genome using tophat.
Canadian Bioinformatics Workshops Ppt Download. At the very end we can compare these results to the results we got from mapping directly to the. RNA-seq Read Mapping with TOPHAT and STAR.
Configure the package specifying the install path and the library dependencies as needed. In this tutorial well use some sample data from a project we did on flies Drosophila melanogaster to illustrate how you can use RNA-seq data to look for differentially expressed genes. Mapping to a reference genome with tophat and counting with HT-seq.
The requirements for aligning this type of data is slightly different from eg. The Bowtie site provides pre-built indices for human mouse fruit fly and others. Background Web Resources.
There are several types of RNA-Seq. Some of the applications used in RNA sequencing analysis are the following. RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Advanced RNA-Seq Analysis topics Hands-on tutorials Analyzing human and potato RNA-Seq data using Tophat and Cufflinks in Galaxy.
This tutorial from 2017 covers the TopHat aligner. It aligns RNA-Seq reads to mammalian-sized genomes using the ultra high-throughput short read aligner Bowtie and then analyzes the mapping results to identify splice junctions between exons. This is quite different conceptually to mapping to the transcriptome directly.
Critically the number of short reads generated for a particular RNA is assumed to be proportional to the. TopHat is a fast splice junction mapper for RNA-Seq reads. If theres no index for your organism its easy to build one yourself.
Jeremy Goecks Galaxy RNAseq tutorial httpmaing2bxpsueduujeremypgalaxy-rna-seq-analysis-exercise. Tophat is a splicing aware aligner so we can map transcripts to the genome. Well try a few different approaches to see whether different tools give similar results.
Transcriptome splice-variantTSSUTR analysis microRNA-Seq etc.
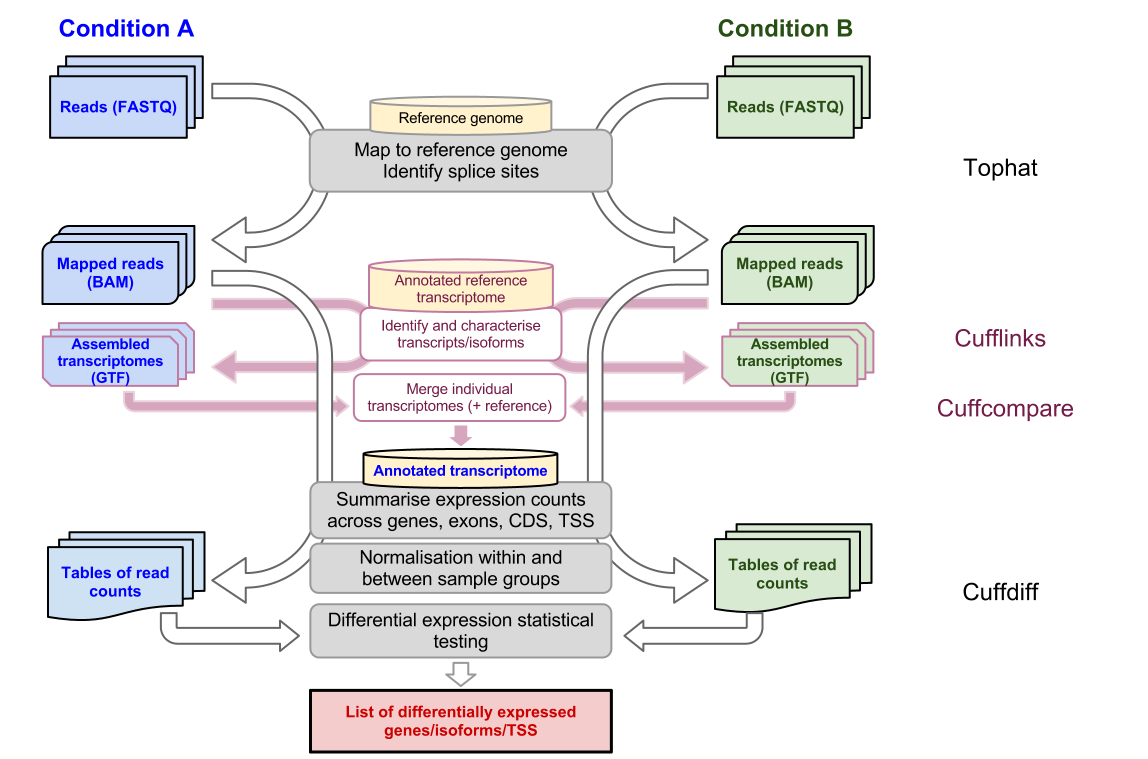
Introduction To Bulk Rnaseq Analysis Bioinformatics Documentation
Basic Analyses With Tophat Cufflinks Rnaseq Tutorial 1 Documentation
Aligning Rna Seq Data Ngs Analysis
The Cufflinks Rna Seq Workflow

Rna Seq Alignment And Visualization Youtube

Reference Based Rnaseq Data Analysis Long
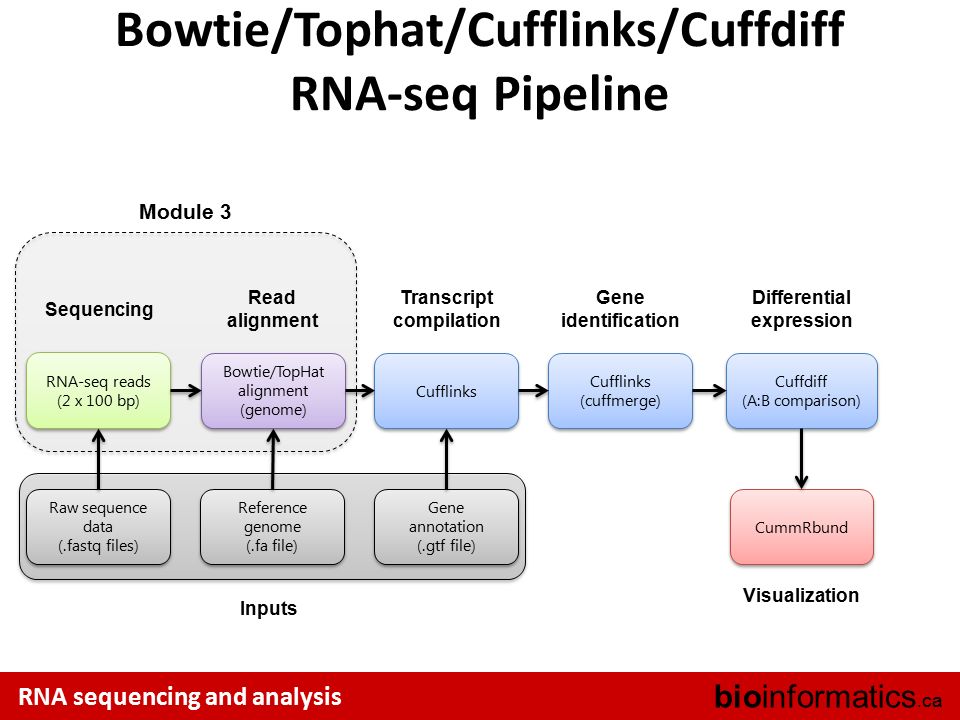
0 komentar
Posting Komentar